Integrating Augmented Reality into bioscience education

Proteins are the highly dynamic molecular machines of the cell – they are often complicated, with their function being dependent on a specific 3-dimentional (3D) structure. This structure-to-function relationship is not often encapsulated effectively when delivered as a 2D image on a presentation slide.
Previously the use of 3D printed tactile objects has helped aid students understanding and promote a greater depth of engagement with the topic (see article link in further reading). After initially investigating the use of virtual reality (VR) headsets to display 3D models in a virtual space, we found that this can be a solitary experience unless the view of the individual wearing the headset is cast to a secondary monitor. As a result, we looked for alternative technologies to bring this topic to life.
In contrast, augmented reality (AR) uses a digital device (such as a phone or tablet) to overlay information onto the environment. The information overlaid can include text, images, videos, or even 3D models mapped in space – allowing the user to move around the object to obtain a different view or even for multiple individuals to view the same object from a different angle.
The augmented content can either be programmed to appear at a specific geographic location or relative to a specific trigger image (which acts a little bit like a bar code). The user’s phone then renders the model relative to the phone and the trigger image to provide a unique perspective of the object which can be shared between multiple devices. Importantly, the advanced power of digital devices (capable of processing complex models and images) has made this technology more accessible and feasible for use in a range of learning environments.
An AR-enhanced learning session can be created by exporting 3D-protein models from the protein databank into pymol (converting .pdb to an object file .obj using ‘> save [filename].obj’ in pymol’s command line). Optional: 3D models can be modified in programmes such as 3DS Max or Blender and saved as .fbx files to highlight domains/peptides or side chains that are functionally important as well as optimizing the model’s performance by adjusting model geometry.
After exploring several platforms capable of generating AR content, ZapWorks was chosen as it comprised both a relatively intuitive editing platform (ZapWorks Studio) which enabled generation of the content including mapping to a trigger image as well as an accompanying mobile phone application (Zappar) which can be used to scan the trigger image and map the 3D structure in real time.
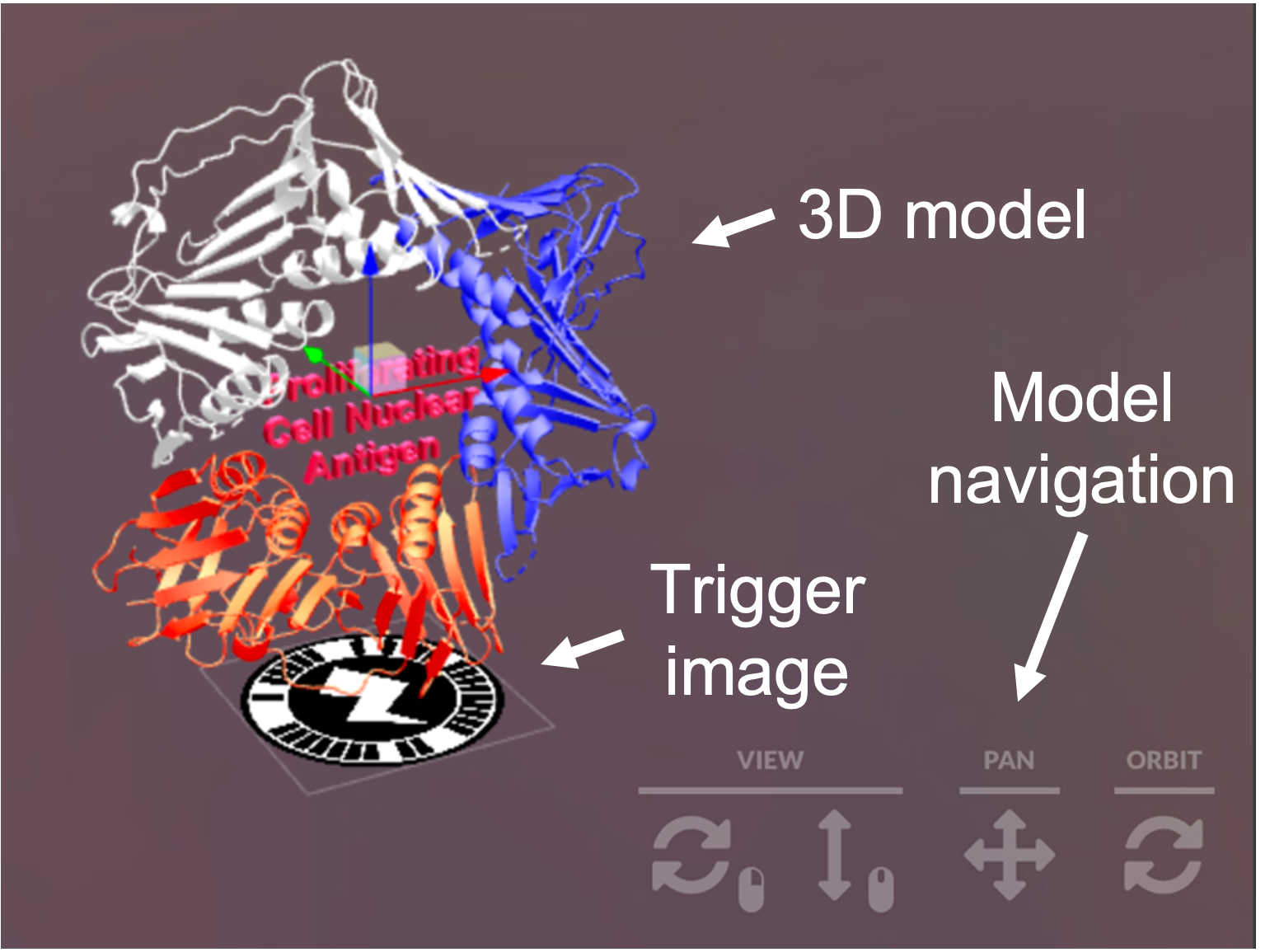
The 3D models (OBJ or FBX) can be imported into ZapWorks studio which allows mapping to trigger images (i.e. the circular barcode which positions the model in space) – other premade models can be imported from an online database called ‘Sketchfab’. Detailed guides on how to get started are provided in the resources section below. Once AR content is published, these 3D models can be viewed using the Zappar mobile phone application. For anyone wanting to use the models generated and used in Reeves et al., 2021, all models are available and freely downloadable from Sketchfab (see resources).


Our research to date has shown AR to be a highly engaging way of delivering bioscience content. An interesting facet to explore in future studies will be the ability to hide additional information in the models, either as part of the model (as done here) or by requiring the user to click on a section of the model that then reveals the in a pop-up box. Importantly, the flexibility and power of this technology has yet to be fully explored and it remains an exciting prospect to capture the dynamic changes in protein function while prompting greater collaboration between students.
Resources:
‘How to Guide’ written by Dr Robbie Baldock hosted on Lecturemotely - https://www.lecturemotely.com/_files/ugd/4b6beb_e5675edd7c854f3cb9ca0fb0b6055807.pdf
Protein Databank - https://www.rcsb.org/
ZapWorks - https://zap.works/
ZapWorks Tutorials and Documentation - https://docs.zap.works/studio/getting-started/
Sketchfab (online database of user created 3D models) - https://sketchfab.com/ (models used in Reeves et al., are accessible and free to download from https://sketchfab.com/RobbieBaldock)
Further reading:
https://journal.alt.ac.uk/index.php/rlt/article/view/2572 - Reeves et al., 2021 ‘Use of augmented reality to aid biosciences education and enrich student experience’.
https://www.youtube.com/watch?v=DU34E4NB-FI #DryLabsRealScience presentation on the use of augmented reality in bioscience teaching.
https://network.febs.org/posts/3d-printing-as-a-cheap-way-of-creating-macromolecular-models-usable-for-teaching - 3D printing models for education FEBs article




Join the FEBS Network today
Joining the FEBS Network’s molecular life sciences community enables you to access special content on the site, present your profile, 'follow' contributors, 'comment' on and 'like' content, post your own content, and set up a tailored email digest for updates.