Embedding authentic research-based learning into a large final-year undergraduate unit

I had the pleasure of attending the inaugural FEBS Molecular Life Sciences Education and Training Conference in Antalya earlier this year. I was also selected to deliver a talk outlining the model for team-based final-year dissertation projects on our transnational joint programme in China. My presentation highlighted the project which exploits the many technical advantages of budding yeast to integrate authentic research into this high enrolment unit.
Investigative Skills
Final-year projects and dissertations are high-impact educational activities that directly engage undergraduates with research (Adams, 2009; Rowley & Slack, 2004; Biggs, 2003; Brew & Boud,1995; Imafuku et al., 2015; Healey, 2023) and encourage deeper learning. They commonly feature in Bioscience degrees, where graduates are expected to gain practical, data analysis and communication skills, in addition to discipline knowledge and awareness of current research (The Quality Assurance Agency for Higher Education, 2023).
Our students on the Nanchang University Joint Programme in Biomedical Sciences (based in China) study for three years toward the Queen Mary Biomedical Sciences degree, followed by two years studying Clinical Medicine at Nanchang University. Completing a major dissertation project in the third year is compulsory, but with the expansion of our cohort size, the traditional apprentice model for dissertation projects was no longer feasible. Investigative Skills is the team-based projects unit that addressed this, where six projects on diverse themes are delivered in dedicated teaching laboratories in Nanchang over a two-week, timetabled research block (Lund & Clyne, 2022). Up to 25 students are allocated to each project theme, subdivided into 4–5 teams. Each team investigates different elements of the overall theme using a variety of biochemical techniques.
Running now for 8 years, Investigative Skills has proven to be a sustainable model for authentic and inclusive dissertation research for large cohorts, where students work in teams to investigate open-ended, suitably challenging research questions using a variety of robust, state-of-the-art techniques. In feedback, more than 80% of respondents agreed that their project involved authentic research, that they had learned the steps required to perform scientific research, and that working in teams made it easier to accomplish the project goals (Lund & Clyne, 2022).
Investigating longevity using a budding yeast model system
The unicellular yeasts are powerful eukaryotic model organisms due to their ease of manipulation and array of available experimental tools, which also makes them advantageous for course-based biochemistry research (Duina et al., 2014; Zhao & Lieberman, 1995; Sherman, 2002; Botstein & Fink, 2011). My collaborator on this project, Professor Gwo-Jiunn Hwang, leads a research group at Nanchang University investigating long non-coding RNAs using budding yeast. Together we designed an Investigative Skills project, based on his ongoing research, that utilises budding yeast to investigate whether the expression and functions of the RPL36 ribosomal protein paralogs are influenced by the overlapping long non-coding RNA genes.
In the budding yeast genome, ~11% of ribosomal protein gene sequences contain overlapping lncRNA gene sequences, which raises a number of interesting mechanistic questions. For example, alteration of ribosomal composition and function might influence the cellular proteome, leading to re-modelling of cellular metabolism and physiology. For example, alteration of ribosomal composition and function might influence the cellular proteome, leading to remodelling of cellular metabolism and physiology. For this undergraduate project, we selected the YMR193C-A and YPL250W-A lncRNA genes, which overlap the RPL36A and RPL36B genes, respectively. Using test strains with genetic modifications that alter lifespan, the main aims of this project for students are (1) to measure RNA and/or protein levels of RPL36A, RPL36B, and the overlapping lncRNA genes during extended culture to investigate reciprocal interactions and (2) to discern any cell cycle progression patterns associated with long-lived strains in comparison to controls (Hwang & Clyne, 2023).
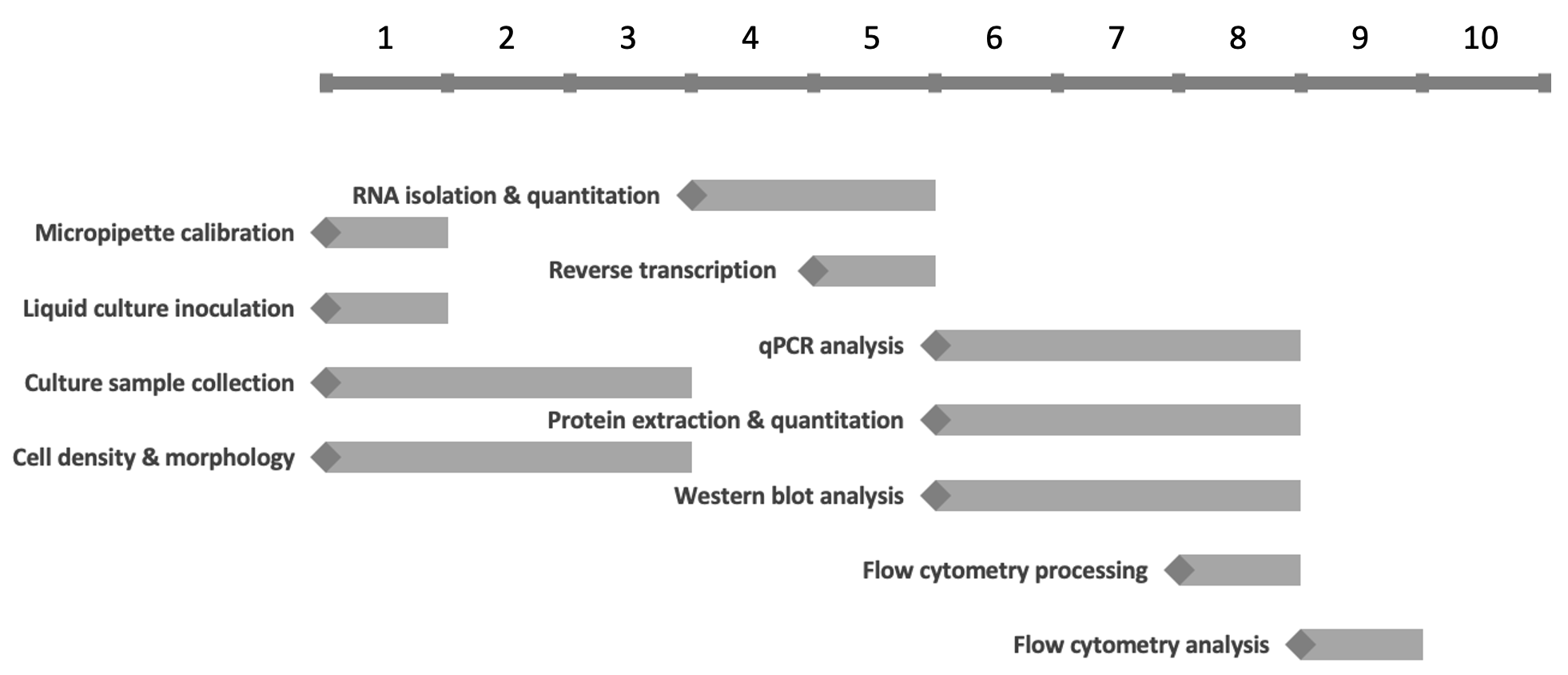
The experimental workplan integrates basic and advanced biochemical techniques, typically including aseptic yeast cell culture and sample collection, RNA isolation and qRT-PCR quantitation, protein extraction and Western blotting, and cell cycle analysis using light microscopy and flow cytometry (Figure 1). Student results have shown that the expression and functions of the RPL36 ribosomal paralogs are indeed influenced by the overlapping lncRNA genes, raising interesting directions for future study.
Embedding authentic research-based learning into curriculum at scale
It is challenging to design research activities that are practical and economical, yet still meaningful. We successfully integrated model organism research into a high-enrolment, final-year undergraduate dissertation unit, where students collaborate on investigating authentic scientific questions with unknown results. In addition to proficiency and increased confidence in a range of experimental skills, such projects illustrate the process of authentic research, immerse students in contemporary research topics, and offer scope for them to contemplate findings and future directions. They can be designed to explore fundamental, conserved biochemical phenomena, while accommodating the demands of large student groups and constraints of course schedules.
Acknowledgements
I thank Professor Gwo-Jiunn Hwang, from Nanchang University, for his collaboration on the design and implementation of this project and Dr. Ying Ying (also at Nanchang University) for her expert practical supervision of this project since its inception. Thanks also to the Nanchang University technical teams who support Investigative Skills, to the Queen Mary supervisors of the lncRNA project, and to our project students for their engagement.
The collaboration resulting in this project began from a chance meeting at a conference. I am very grateful for having the opportunity to present our pedagogical work in Antalya and engage with so many enthusiastic educators in this wonderful FEBS community. I would also like the thank the FEBS Education & Training Committee for the travel grant award, which enabled me to attend.
References
- Adams DJ. 2009. Current trends in laboratory class teaching in university bioscience programmes. Biosci. Educ. 13: 1–14. doi: 10.3108/beej.13.3.
- Biggs J. 2003. Teaching for Quality Learning at University. Buckingham: Open University Press.
- Botstein D, Fink GR. 2011. Yeast: an experimental organism for 21st Century biology. Genetics 189(3): 695-704. doi: 10.1534/genetics.111.130765.
- Brew A, Boud D. 1995. Teaching and research: establishing the vital link with learning. Higher Education 29: 261–273. doi: 10.1007/BF01384493.
- Duina AA, Miller ME, Keeney JB. 2014. Budding yeast for budding geneticists: a primer on the Saccharomyces cerevisiae model system. Genetics 197(1):33-48. doi: 10.1534/genetics.114.163188.
- Healey M. Refreshing the curriculum: approaches to curriculum design. Available from: www.healeyheconsultants.co.uk/resources [Accessed on 10th June 2023]
- Hwang GJ and Clyne RK. 2023. Long non-coding RNA and ribosomal protein genes in a yeast ageing model: an investigation for undergraduate research-based learning. Essays in Biochemistry 67(5): 893-901.
- Imafuku R, Saiki T, Kawakami C, Suzuki Y. 2015. How do students’ perceptions of research and approaches to learning change in undergraduate research? Int J Med Educ 6: 47–55. doi: 10.5116/ijme.5523.2b9e.
- Lund T and Clyne RK. 2022. Implementation and evaluation of a team-based authentic research project module for large cohorts. Essays in Biochemistry 66(1): 45-51.
- The Quality Assurance Agency for Higher Education. Subject Benchmark Statement Biosciences. Available at: www.qaa.ac.uk/docs/qaa/sbs/sbs-biosciences-23.pdf?sfvrsn=b570a881_4 [Accessed 10th June 2023].
- Rowley J, Slack F. 2004. What is the future for undergraduate dissertations? Educ Train 246: 176–181. doi: 10.1108/00400910410543964.
- Sherman F. 2002. Getting started with yeast. Methods Enzymol 350: 3-41. doi: 10.1016/s0076-6879(02)50954-x.
- Zhao Y, Lieberman HB. 1995. Schizosaccharomyces pombe: a model for molecular studies of eukaryotic genes. DNA Cell Biol 14(5): 359-71. doi: 10.1089/dna.1995.14.359.
Image by Donnie0102 from Pixabay.




Join the FEBS Network today
Joining the FEBS Network’s molecular life sciences community enables you to access special content on the site, present your profile, 'follow' contributors, 'comment' on and 'like' content, post your own content, and set up a tailored email digest for updates.