The biochemistry of SARS-CoV-2: a reasoned collection of resources

This post presents a reasoned collection of websites and papers on the biochemistry of SARS-CoV2. The selection is unavoidably arbitrary, but the post will be periodically updated and extended. If you have suggestions, you are most welcome to collaborate and to provide the necessary links in the comment section.
Hubs and Data Banks
- The RCSB Protein Data Bank website has a webpage on structural biology research on Coronavirus.
- The UniProt website has a webpage that reports the annotated sequences of proteins from Covid-19.
- GenBank has a page dedicated to SARS-CoV-2. Moreover you can access the NCBI virus index
- BLAST has a resource to investigate and compare all nucleotide sequences from betacoronaviruses
Articles and resources on the viral genome: 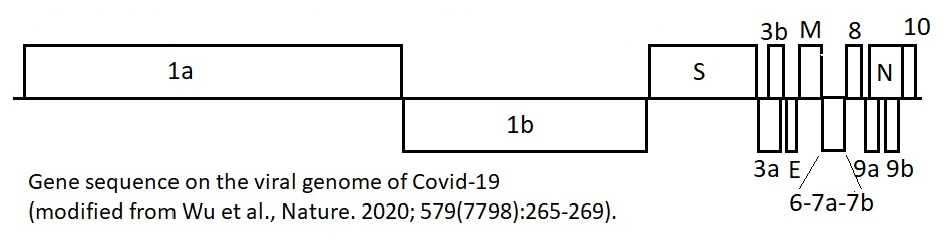
- Lu R, Zhao X, Li J, Niu P, Yang B, Wu H, Wang W, Song H, Huang B, Zhu N, Bi Y, Ma X, Zhan F, Wang L, Hu T, Zhou H, Hu Z, Zhou W, Zhao L, Chen J, Meng Y, Wang J, Lin Y, Yuan J, Xie Z, Ma J, Liu WJ, Wang D, Xu W, Holmes EC, Gao GF, Wu G, Chen W, Shi W, Tan W. Genomic characterisation and epidemiology of 2019 novel coronavirus: implications for virus origins and receptor binding. Lancet. 2020 Feb 22;395(10224):565-574. doi: 10.1016/S0140-6736(20)30251-8. Epub 2020 Jan 30.
- Wu F, Zhao S, Yu B, Chen YM, Wang W, Song ZG, Hu Y, Tao ZW, Tian JH, Pei YY, Yuan ML, Zhang YL, Dai FH, Liu Y, Wang QM, Zheng JJ, Xu L, Holmes EC, Zhang YZ. A new coronavirus associated with human respiratory disease in China. Nature. 2020 Mar;579(7798):265-269. doi: 10.1038/s41586-020-2008-3.
- Mousavizadeh L, Ghasemi S. Genotype and phenotype of COVID-19: Their roles in pathogenesis. J Microbiol Immunol Infect. 2020 Mar 31. pii: S1684-1182(20)30082-7. doi: 10.1016/j.jmii.2020.03.022.
- Forster P, Forster L, Renfrew C, Forster M. Phylogenetic network analysis of SARS-CoV-2 genomes. Proc Natl Acad Sci U S A. 2020; pii: 202004999. doi: 10.1073/pnas.2004999117.
Journal articles and pre-prints on the viral proteases:
- Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020 Mar 20. pii: eabb3405. doi: 10.1126/science.abb3405. Structure of and binding of specific inhibitors to the main protease from Covid-19. This protease is a symmetric homodimer, whose catalytic residues are His41 and Cys145. The two catalytic sites are on oposite sides of the dimer, far from one another and from the monomer-monomer interface.
- Wang L, Hu W, Fan C. Structural and biochemical characterization of SADS-CoV papain-like protease 2. Protein Sci. 2020 Mar 26. doi: 10.1002/pro.3857. Processing of the polyproteins pp1a and pp1ab produced by coronaviruses requires at least two or three specific viral proteases. This paper reports the structure and properties of the papain-like protease from a swine coronavirus related to Covid-19.
- Zhang L, Lin D, Sun X, Curth U, Drosten C, Sauerhering L, Becker S, Rox K, Hilgenfeld R. Crystal structure of SARS-CoV-2 main protease provides a basis for design of improved α-ketoamide inhibitors. Science. 2020; 368: 409-412. doi: 10.1126/science.abb3405.
Journal articles and pre-prints on the spike protein and its receptor (ACE2):
- Lan J, Ge J, Yu J, Shan S, Zhou H, Fan S, Zhang Q, Shi X, Wang Q, Zhang L, Wang X. Structure of the SARS-CoV-2 spike receptor-binding domain bound to the ACE2 receptor. Nature. 2020 Mar 30. doi: 0.1038/s41586-020-2180-5.
- Wrapp D, Wang N, Corbett KS, Goldsmith JA, Hsieh CL, Abiona O, Graham BS, McLellan JS. Cryo-EM structure of the 2019-nCoV spike in the prefusion conformation. Science. 2020 Mar 13;367(6483):1260-1263. doi: 10.1126/science.abb2507.
- Mallapaty S. Why does the coronavirus spread so easily between people? Nature. 2020 Mar;579(7798):183. doi: 10.1038/d41586-020-00660-x. A summary of the biochemical mechanisms of Covid-19 binding to ACE2 on the host cell membrane and its proteolytic activation by furin, which explains the broad organ specificity of the virus.
- Hussain M, Jabeen N, Raza F, Shabbir S, Baig AA, Amanullah A, Aziz B. Structural Variations in Human ACE2 may Influence its Binding with SARS-CoV-2 Spike Protein. J Med Virol. 2020 Apr 6. doi: 10.1002/jmv.25832. Genetic variants of ACE2, the human protein that acts as the receptor for Covid-19, may confer resistance to the infection. This has important implications for the predictions of the possible attack rate of the pandemics and the public policy decisions on its containment.
- Ou X, Liu Y, Lei X, Li P, Mi D, Ren L, Guo L, Guo R, Chen T, Hu J, Xiang Z, Mu Z, Chen X, Chen J, Hu K, Jin Q, Wang J, Qian Z Characterization of spike glycoprotein of SARS-CoV-2 on virus entry and its immune cross-reactivity with SARS-CoV. Nat Commun. 2020 Mar 27;11(1):1620. doi: 10.1038/s41467-020-15562-9.
- Shang J, Ye G, Shi K, Wan Y, Luo C, Aihara H, Geng Q, Auerbach A, Li F. Structural basis of receptor recognition by SARS-CoV-2. Nature. 2020 Mar 30. doi: 10.1038/s41586-020-2179-y. PMID: 32225175
- Yan R, Zhang Y, Li Y, Xia L, Guo Y, Zhou Q. Structural basis for the recognition of SARS-CoV-2 by full-length human ACE2. Science. 2020 Mar 27;367(6485):1444-1448. doi: 10.1126/science.abb2762.
Journal articles and pre-prints on the viral RNA-dependent RNA polymerase and endonuclease(s):
- Yan Gao, Liming Yan, Yucen Huang, Fengjiang Liu, Yao Zhao, Lin Cao, Tao Wang, Qianqian Sun, Zhenhua Ming, Lianqi Zhang, Ji Ge, Litao Zheng, Ying Zhang, Haofeng Wang, Yan Zhu, Chen Zhu, Tianyu Hu, Tian Hua, Bing Zhang, Xiuna Yang, Jun Li, Haitao Yang, Zhijie Liu, Wenqing Xu, Luke W. Guddat, Quan Wang, Zhiyong Lou, Zihe Rao Structure of RNA-dependent RNA polymerase from 2019-nCoV, a major antiviral drug targetbioRXiv, doi: https://doi.org/10.1101/2020.03.16.993386. This paper reports the cryo-EM structure of 2019-nCoV full-length RNA-dependent RNA polymerase (protein nsp12) in complex with cofactors nsp7 and nsp8 at a resolution of 2.9 A.
- Gordon CJ, Tchesnokov EP, Feng JY, Porter DP, Gotte M. The antiviral compound remdesivir potently inhibits RNA-dependent RNA polymerase from Middle East respiratory syndrome coronavirus. J Biol Chem. 2020 Feb 24. pii: jbc.AC120.013056. doi: 10.1074/jbc.AC120.013056. Remdesvir is synthetic nucleotide analog and a selective inhibitor of viral RNA-dependent RNA polymerases. This paper demonstrates that it inhibitits the replication of the MERS Covid, a virus species related to Covid-19.
- Youngchang Kim, Robert Jedrzejczak, Natalia I. Maltseva, Michael Endres, Adam Godzik, Karolina Michalska, Andrzej Joachimiak Crystal structure of Nsp15 endoribonuclease NendoU from SARS-CoV-2 bioRXiv doi: https://doi.org/10.1101/2020.03.02.968388
Journal articles and pre-prints on the nucleocapsid protein (N):
- Tsung-Lin Tsai, Ching-Houng Lin, Chao-Nan Lin, Chen-Yu Lo, Hung-Yi Wu Interplay between the Poly(A) Tail, Poly(A)-Binding Protein, and Coronavirus Nucleocapsid Protein Regulates Gene Expression of Coronavirus and the Host Cell. J. Virol.2018; 92:e01162-18. Although this is not the N protein from SARS-CoV2, but from a related bovine coronavirus, it is the only paper I found reporting this peculiar behaviour of th N protein.
Page updated April 29, 2020.




Join the FEBS Network today
Joining the FEBS Network’s molecular life sciences community enables you to access special content on the site, present your profile, 'follow' contributors, 'comment' on and 'like' content, post your own content, and set up a tailored email digest for updates.
Dear Andrea,
I have seen a nice publication in the recent issue of Science from the group of Rolf Hingenfeld who is excellent strucural biologist working in Lübeck.
Crystal Strucutre of SARC-COV-2 main protease provides a basis for design of improves alfa.ketoamide inhibitors. Science 24 April 2020 Vol 368 Issue 6489 pp 409-412 open access
Thank you Laszlo, I'm adding it to the list! Andrea
Dear Andrea,
Another important development.
Laszlo
A SARS-CoV-2 protein interaction map reveals targets for drug repurposing David E. Gordon, Gwendolyn M. Jang, […]Nevan J. Krogan
https://www.nature.com/articles/s41586-020-2286-9