The FEBS Journal Open Access Round-up

On your Mark(er)s, get set, go?: The hidden effects of auxotrophic markers on yeast phenotypes 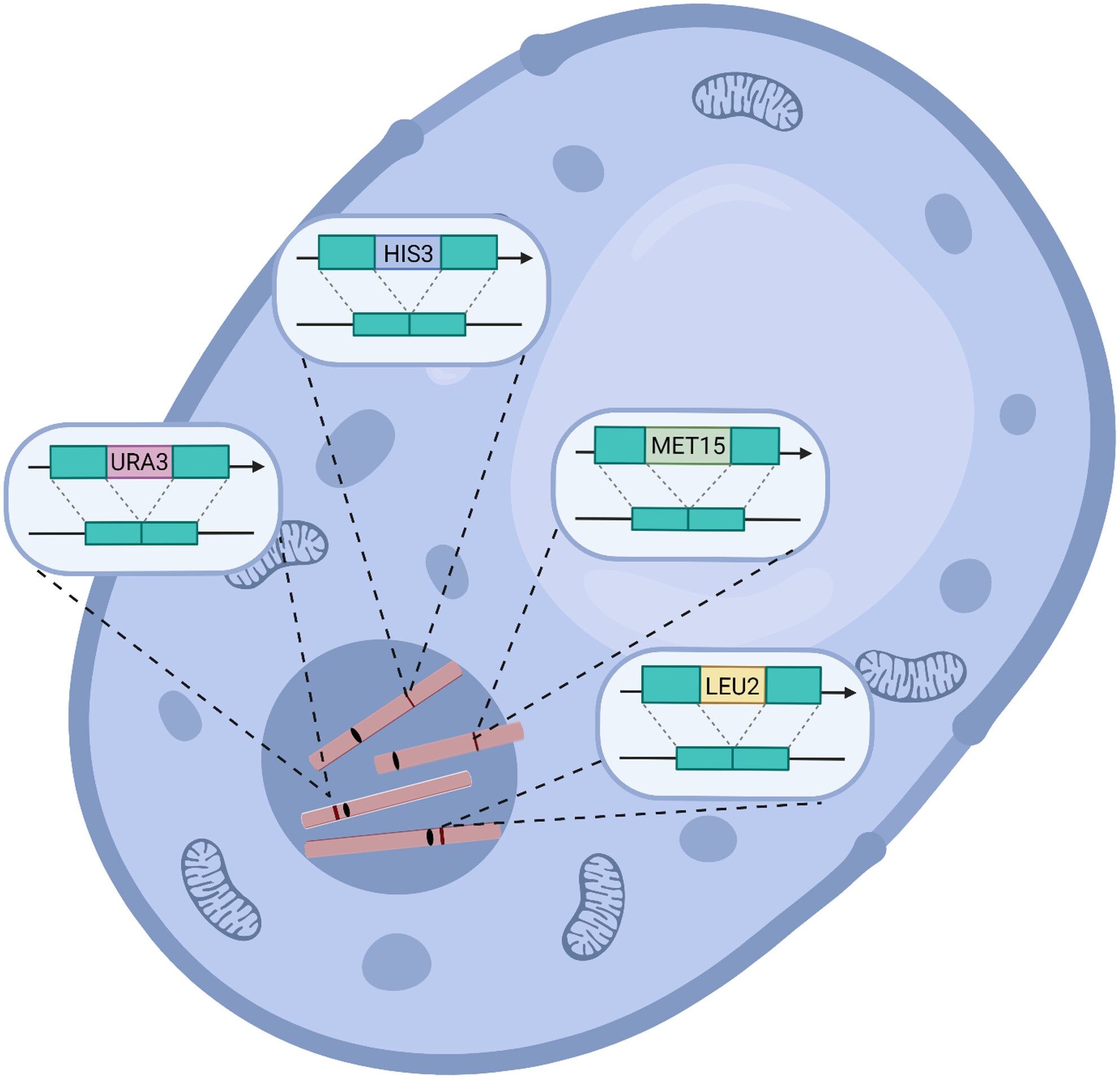
Attention budding scientist! A new study by Kaplan, Levkovich and colleagues has put the common auxotrophic markers from the frequently used Saccharomyces cerevisiae strain BY4741 under the spotlight. This “wild-type” auxotrophic strain of yeast carries deletions in four essential genes in the biosynthesis of leucine, uracil, histidine and methionine. These markers are fundamental tools in yeast genetic engineering, allowing us researchers to easily select and manipulate yeast cells by using synthetic growth media that lack the required metabolites. The central question asked by the authors was: do these markers have any unintended consequences for the yeast phenotypes? (spoiler: yes. But don’t stop reading). To answer this question, they generated knock-in yeast strains that contained all possible combinations of the four auxotrophic markers, leu2∆0, ura3∆0, his3∆1, met15∆0, and a prototrophic knock-in. Using the cellular stressors rapamycin, acetic acid, and ethanol, Kaplan and team found that leucine auxotrophy has the biggest impact on phenotype. Focusing next on rapamycin sensitivity. They discovered that many mutants that were previously annotated as rapamycin-sensitive were influenced by the auxotrophic markers, and that their sensitivity was reduced or eliminated in the prototrophic background. These results highlight a significant phenotypic bias caused by common auxotrophic markers and that the yeast research community should carefully consider the metabolic background of laboratory yeast strains .
NIPP1: going full circle on PP1 Activation
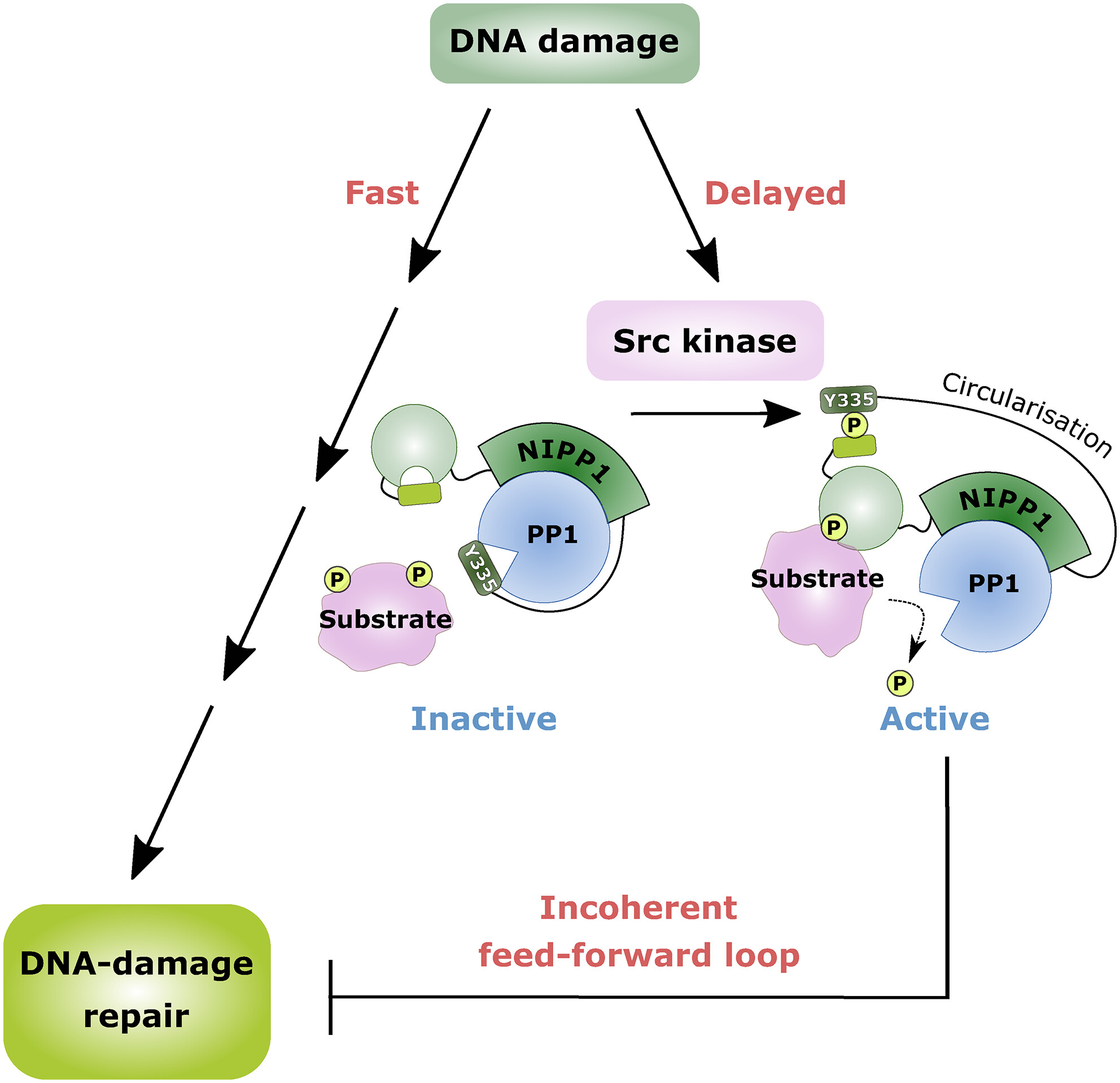
When you think of repairs, they don’t come much more important to life than the accurate and efficient repair of DNA following damage. The delayed or timely response to DNA damage is the focus of an exciting new paper by Wu et al. Here, the authors focus on the activation of the PP1:NIPP1 holoenzyme. PP1 is a Ser/Thr phosphatase, while NIPP1 is one of its inhibitors. NIPP1 has an intrinsically disordered PP1-anchoring domain with short linear motifs (SLiMs) that mediate binding to surface grooves of PP1 along with an N-terminal substrate-recruiting FHA domain, and a C-terminal PP1-inhibitory domain. Under normal conditions, the PP1:NIPP1 is inactive , so it needs to be turned on by DNA damage. Building upon previous knowledge that NIPP1 is phosphorylated at the C terminus by Src Kinase, Wu and colleagues showed that this phosphorylation peaks several hours after UV-induced DNA damage. This has two implications; it disinhibits PP1 and allows NIPP1 to form a circular structure that helps it bring more substrates to PP1. The authors also showed that constitutively activating PP1 and NIPP1 via a phosphomimetic mutation at the C-terminal domain impairs DNA repair and makes cells more sensitive to DNA damage. This shows how essential a precisely orchestrated DNA repair cascade is for the cell, and this knowledge could pave the way to more efficacious DNA-damaging agents for cancer therapy.
Cryo-EM captures the primosome in action
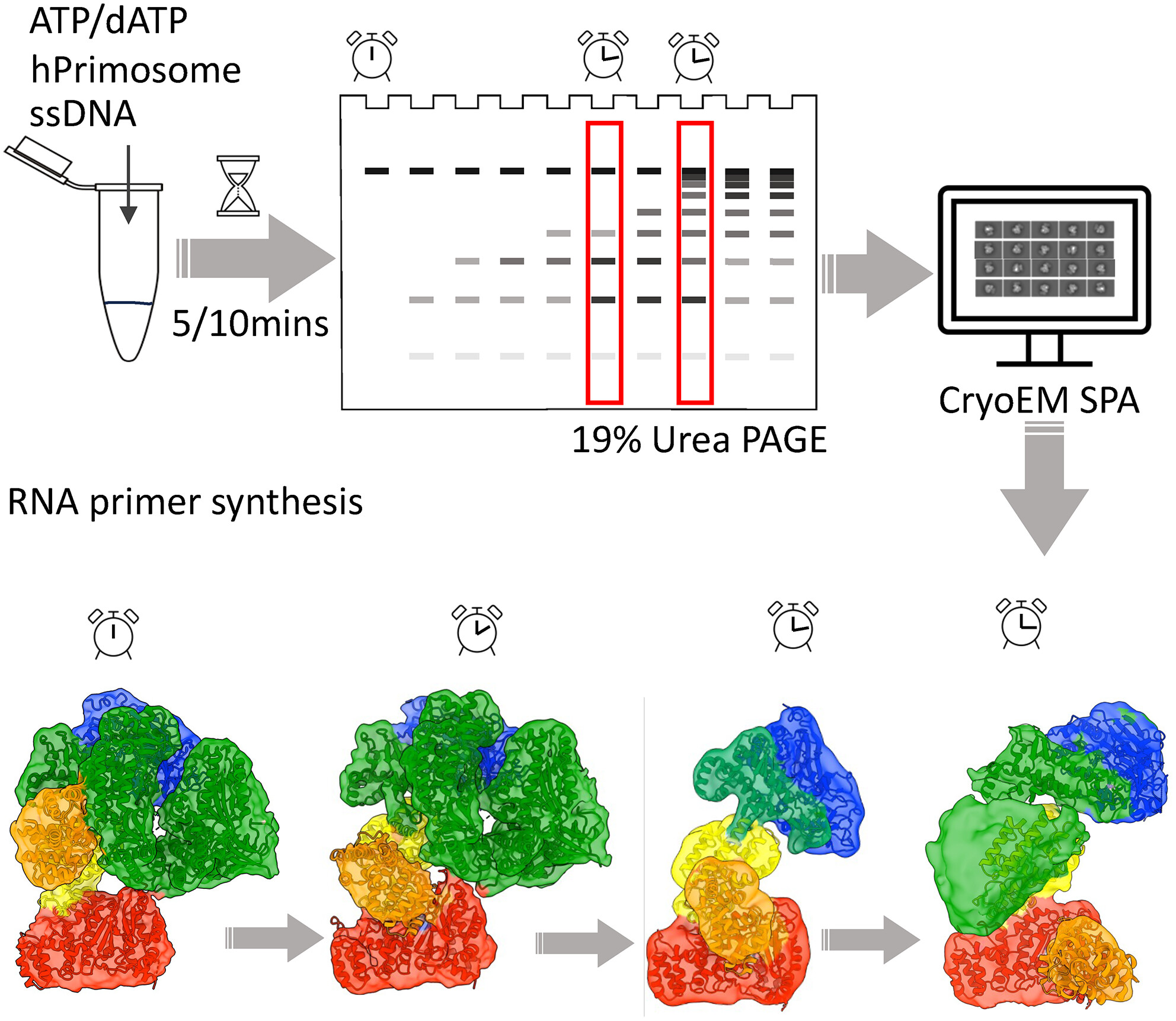
In all kingdoms of life, DNA replication requires the production of RNA-DNA hybrid primers. These are created by the primosome, a heterotetrametric complex composed of primase and polymerase alpha (Pol α). Primase synthesises short RNA primers that are handed over to Pol α, which extends them to generate 20-30nt hybrid RNA-DNA primers. To solve this complex yet poorly understood choreography, Yin and colleagues used Cryo-EM to unravel how primase kickstarts primer generation and transfers the primer to the catalytic domain of pol α (POLA1). The authors identified four conformational intermediates of the human primosome as it transitions from the apo-state to one that is competent for initiation and subsequent handover of the RNA primer to POLA1. These mechanistic insights reveal how the primase subunits PRIM1 and PRIM2 are held apart in a compact auto-inhibitory state, and how the binding of an NTP and ssDNA then unlock PRIM1 and allow it to dock to PRIM2’s active site. This releases the POLA1 and initiates the synthesis of the RNA primer. The authors also showed that PRIM2 tracks the 5′ end of the growing RNA primer and that POLA1 interacts with PRIM2 to regulate primer size and termination in a dATP-dependent manner. This study fills a critical gap in our understanding of how DNA replication initiates. It also has implications for human health, as defects in the primosome can cause genetic diseases and cancer.
*The FEBS Journal is a multidisciplinary, non-profit society journal that publishes full-length high-quality papers and expert reviews relevant to the molecular, cellular and biochemical life sciences. Send us your next paper, it will be in great company!
References
-
Kaplan, K., Levkovich, S.A., DeRowe, Y., Gazit, E. and Laor Bar-Yosef, D. (2024), Mind your marker: the effect of common auxotrophic markers on complex traits in yeast. FEBS J. https://doi.org/10.1111/febs.17095
-
Wu, D., Van der Hoeven, G., Claes, Z., Van Eynde, A. and Bollen, M. (2024), DNA damage-induced allosteric activation of protein phosphatase PP1:NIPP1 through Src kinase-induced circularization of NIPP1. FEBS J. https://doi.org/10.1111/febs.17064
-
Yin, Z., Kilkenny, M.L., Ker, D.-S. and Pellegrini, L. (2024), CryoEM insights into RNA primer synthesis by the human primosome. FEBS J. https://doi.org/10.1111/febs.17082




Join the FEBS Network today
Joining the FEBS Network’s molecular life sciences community enables you to access special content on the site, present your profile, 'follow' contributors, 'comment' on and 'like' content, post your own content, and set up a tailored email digest for updates.