The FEBS Journal Open Access Round-up #4

Get ready for a PPEP talk.
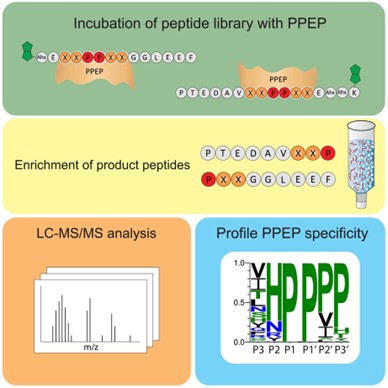
Bacterial proteases are a diverse class of enzymes and are of key importance in bacterial pathogenesis; as antimicrobial drug targets and in the wider biotechnological industry. Pro-Pro Endopeptidases (PPEPs) stand out amongst their peers for their unique ability to hydrolyse proline-proline bonds within protein structures. This remarkable specificity not only distinguishes PPEPs from other proteases, but also underscores the importance of gaining a comprehensive understanding of their substrate specificity and therefore unlocking their potential for biotechnological applications.
Claushuis et al. set out to do exactly this: by employing synthetic combinatorial peptide libraries alongside mass spectrometry, FRET and structural modelling, they elucidated the precise mechanisms that govern substrate specificity on both the prime and non-prime sides. This approach revealed the optimal substrates of different PPEP variants, such as a new PPEP (termed PPEP-4) isolated from Anoxybacillus tepidamans.
This study establishes a robust method for understanding the specificity of putative PPEPs and is vital for grasping the nuances of bacterial physiology and pathogenesis. Moreover, the insights gained could pave the way for innovative solutions in the pharmaceutical industry, where targeted proteolysis can be a powerful tool for drug development.
Acid Test: The DCA-tecting Revolution in Biosensors
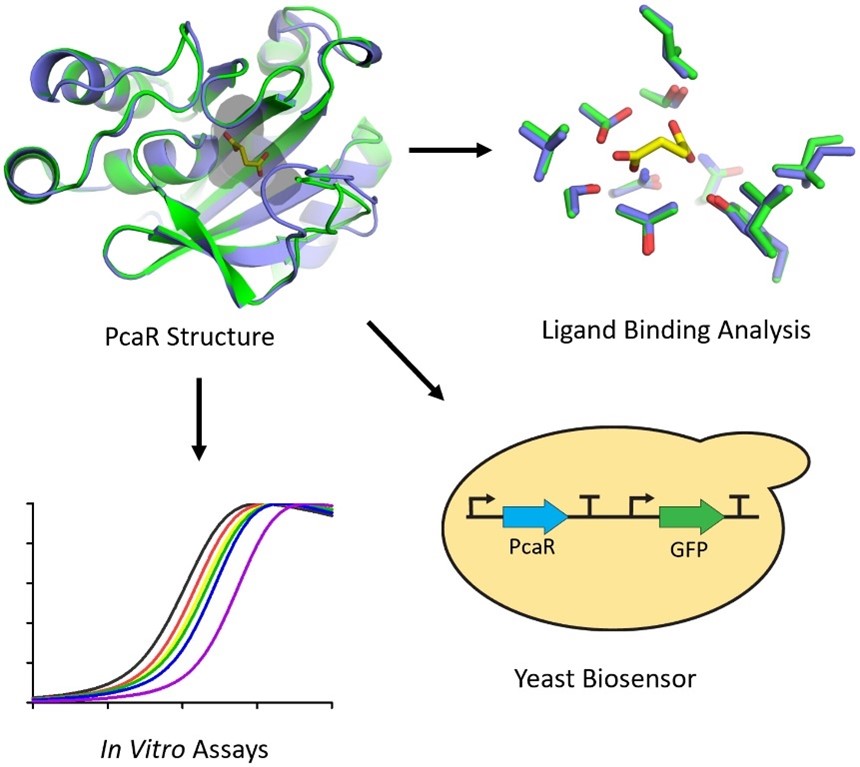
You may not have heard too much about Dicarboxylic acids (DCAs), but they are everywhere, from health and pharmaceuticals to food and textiles. As a result, identifying and detecting DCAs holds significant value for their use in medical applications, environmental stewardship, and processing of industrial products. Prokaryotic transcription factors, which respond to small molecules like DCAs, have therefore emerged as promising biosensors.
In their latest work, Pham et al. have focused on characterising the Pca regulon regulatory protein PcaR, an IcIR transctription factor from the bacterium Pseudomonas putida, as a biosensor for DCAs. PcaR was able to recognise a broad range of DCA ligands, including fumarate and succinate. Next, they investigated the broader applicability of PcaR as a biosensor, extending its base beyond traditional prokaryotic systems. By introducing it into the yeast S. cerevisiae, they demonstrated its capacity to detect multiple DCAs utilising green fluorescence as the readout of the yeast-based DCA sensor. Finally, the authors structurally characterised the PcaR ligand recognition mechanisms for the first time by solving PcaR crystal structures in the succinate-bound and unbound states, unveiling new information about its specificity for binding DCAs. Understanding these mechanisms is essential for engineering PcaR-based biosensors that are both sensitive and selective.
The implications of this study are significant. The potential applications of PcaR-based biosensors are broad, ranging from environmental monitoring to healthcare diagnostics.
*The FEBS Journal is a multidisciplinary, non-profit society journal that publishes full-length high-quality papers and expert reviews relevant to the molecular, cellular and biochemical life sciences. Send us your next paper, it will be in great company!




Join the FEBS Network today
Joining the FEBS Network’s molecular life sciences community enables you to access special content on the site, present your profile, 'follow' contributors, 'comment' on and 'like' content, post your own content, and set up a tailored email digest for updates.