CancerModels.Org – an open global research platform for patient-derived cancer models
What are PDCMs and why are they important?
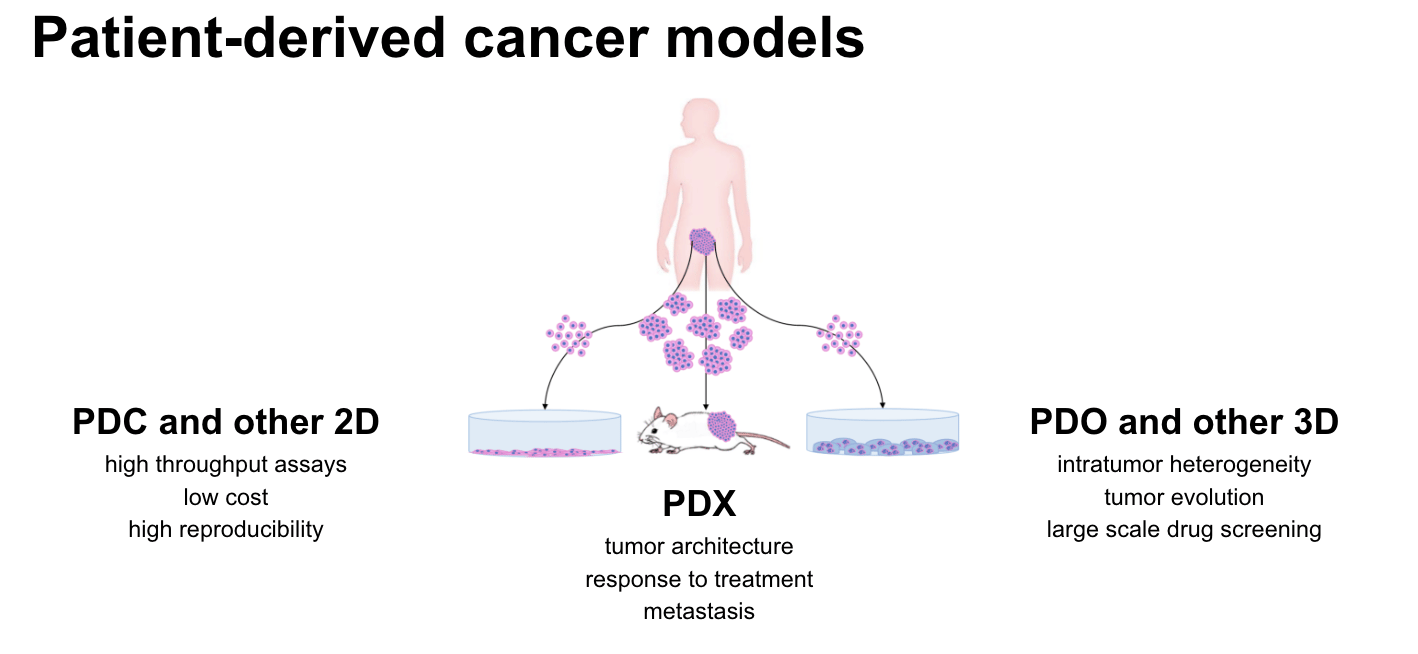
Patient-derived cancer models (PDCMs) are created from tumor samples belonging to individual patients, to mimic as close as possible the underlying characteristics of the patient's cancer. They are a powerful oncology research platform to study cancer progression, mechanisms of drug resistance and predicting response to anti-cancer therapeutic compounds. In principle, there are three major types of models (Figure 1) - patient-derived xenografts (PDX), organoids and cell lines - and each type offers unique advantages tailored to specific research areas (1, 2, 3). Cell lines are cost-effective and support high-throughput assays (4, 5), organoids replicate intratumor heterogeneity and drug response dynamics (6, 7), while patient-derived xenografts (PDXs) retain tumor architecture for better treatment response prediction (8, 9).
These models are also used in functional precision oncology to test the response of the tumor cells to various drugs, allowing for personalized treatment strategies based on the unique biology of each patient's cancer (10). By directly perturbing live tumor cells from patients with drugs, functional precision oncology aims to identify the most effective therapies for each individual, overcoming the limitations of genomics-based precision medicine and enhancing treatment efficacy through personalized and translatable information derived from PDCMs. The models, hence, serve as valuable platforms for anticancer drug testing, co-clinical trials, and the development of personalized treatment approaches tailored to individual patients(11,12).
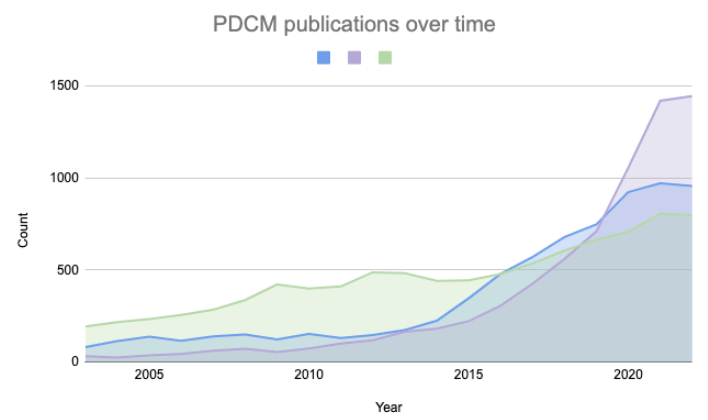
PDCMs are being generated and distributed by many academic groups, research institutes, and commercial organizations and the number of publications mentioning these models has been rapidly growing in the past decade (Figure 2). The heterogeneity of the underlying metadata and the lack of robust standards to describe these models make it difficult for researchers to find models of interest and compare associated data across multiple academic and commercial sources. Finding a model with certain characteristics such as specific mutation or drug response requires one to search through multiple databases and repositories, download the metadata and data from multiple sources often in different formats, transform and analyse it. This is a time-consuming exercise before even starting to test hypotheses and addressing scientific questions. CancerModels.Org addresses the challenge of navigating this fragmented landscape by providing seamless access to PDCMs through its standardized approach.
How CancerModels.Org can help
CancerModels.Org builds bridges across the fragmented PDCM landscape by standardising, harmonising and integrating the complex and diverse data associated with PDCMs using FAIR as foundational principles. It’s main significance is in providing a unified point for PDCM stakeholders, from basic and clinical researchers to bioinformaticians and tool developers, to search and compare over 8300 PDCMs and associated data, including frequently mutated genes, diagnoses, drug treatments and sequence data from PDXs, organoids, and cell lines. It covers multiple cancer types and precancerous conditions, including rare paediatric models and models of ethnic minority backgrounds, making it the largest free-to-consumer and open-access resource of its kind.
Users can search for models via a web interface or the REST API and explore molecular data summaries for models of specific cancer types. The data types include gene expression, gene mutation, copy number alterations, biomarkers, imaging, patient treatment, and drug dosing studies. All data is also available via cBioPortal for visualisation. Moreover, the knowledge is enriched with links to external resources – publication platforms, cancer-specific annotation tools (e.g. COSMIC, CIViC, ClinGen, OncoMX, OpenCRAVAT), and raw data archives (EGA, ENA, dbGAP, GEO, BioImage archive).
Video 1. How to find breast cancer models with mutated BRCA1 and BRCA2 in CancerModels.Org. Watch directly on YouTube.
PDCM Finder project
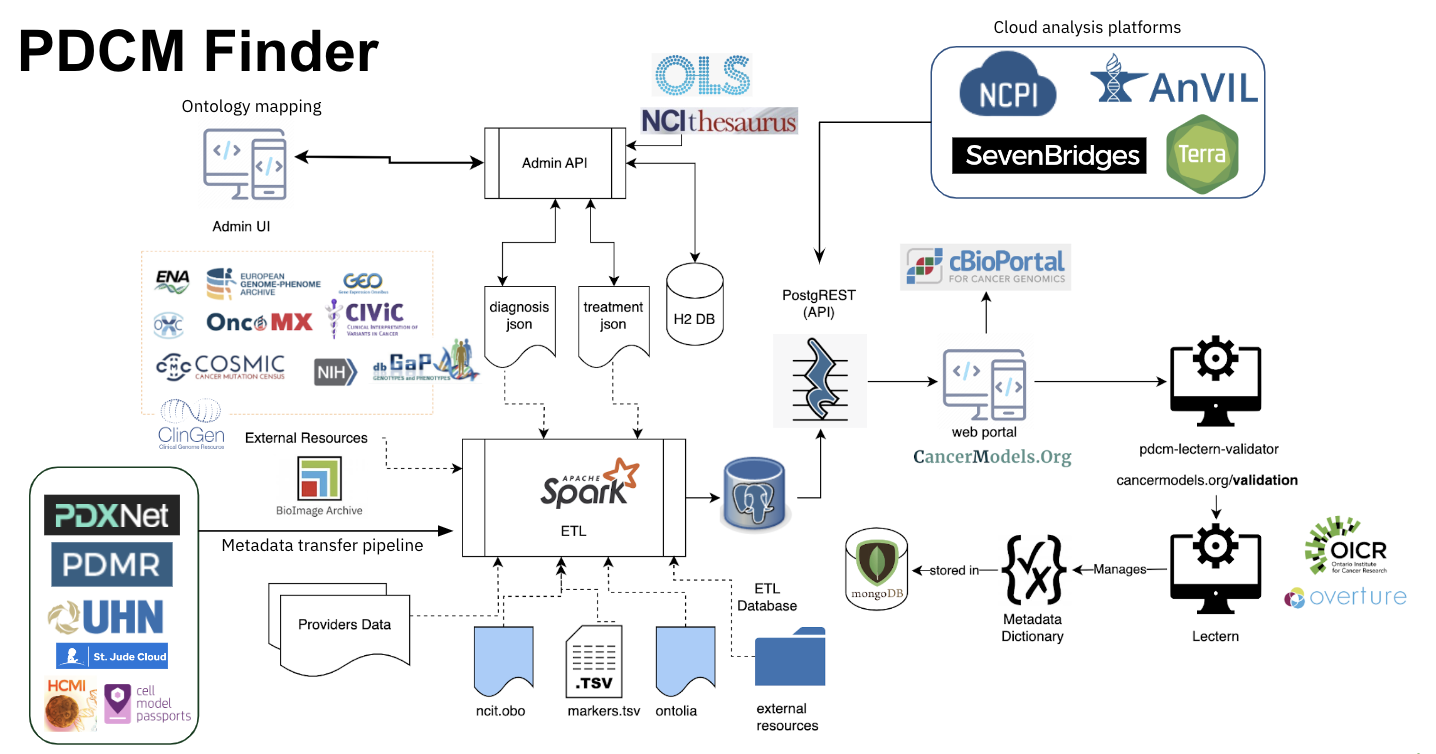
CancerModels.Org platform is a part of the PDCM Finder project that is driving the development of and promoting the use of descriptive standards to facilitate data interoperability and promote global sharing of models. In 2017 we published PDX-MI (14) – a minimal information standard for PDXs developed by the community – and now we are working on the publication of the descriptive standard for the in-vitro PDCMs (manuscript in preparation). We provide expertise and software components to support several worldwide consortia including PDXNet and EurOPDX. The tools and solutions we develop are freely available as a part of the PDCM Finder project under an Apache 2.0 licence.
We develop our resource with a strong emphasis on user-centered design ensuring our users can easily navigate CancerModels.Org. Users can request features and ask questions via our Contact form or provide feedback on any page of the resource following the feedback icon on the lower right corner.
Video 2. New features in CancerModels.Org: Page tour, Feedback bubble and Contact form. Watch directly on YouTube.
We provide a number of training materials in various formats - from the Webinar recorded with the EMBL-EBI training team and downloadable CancerModels.Org Training guide to the Youtube tutorials and the short video updates of new features on LinkedIn and X (former Twitter).
CancerModels.Org is open-source and community-driven hence enabling researchers from various backgrounds and institutions to access and contribute to the shared knowledge pool and increase visibility and reusability of their models. This fosters collaboration across disciplines and geographies, breaking down traditional barriers to information and supporting the broader goals of global health equity in addition to accelerating cancer research and developing personalised treatments.
CancerModels.Org is co-developed by the EMBL-EBI and The Jackson Laboratory and is supported by the National Institutes of Health/National Cancer Institute grants U24 CA204781, U24 CA253539 and R01 CA089713.
References:
- Hou, X., Du, C., Lu, L. et al. Opportunities and challenges of patient-derived models in cancer research: patient-derived xenografts, patient-derived organoid and patient-derived cells. World J Surg Onc 20, 37 (2022). https://doi.org/10.1186/s12957-022-02510-8
- Yoshida GJ. Applications of patient-derived tumor xenograft models and tumor organoids. J Hematol Oncol. 2020;13(1):4. Published 2020 Jan 7. https://doi.org/10.1186/s13045-019-0829-z
- Liu, Y., Wu, W., Cai, C. et al. Patient-derived xenograft models in cancer therapy: technologies and applications. Sig Transduct Target Ther 8, 160 (2023). https://doi.org/10.1038/s41392-023-01419-2
- Jaaks, P., Coker, E.A., Vis, D.J. et al. Effective drug combinations in breast, colon and pancreatic cancer cells. Nature 603, 166–173 (2022). https://doi.org/10.1038/s41586-022-04437-2
- Kim, SY., Lee, J.Y., Kim, D.H. et al. Patient-Derived Cells to Guide Targeted Therapy for Advanced Lung Adenocarcinoma. Sci Rep 9, 19909 (2019). https://doi.org/10.1038/s41598-019-56356-4
- Sato, T., Vries, R., Snippert, H. et al. Single Lgr5 stem cells build crypt-villus structures in vitro without a mesenchymal niche. Nature 459, 262–265 (2009). https://doi.org/10.1038/nature07935
- Verduin Maikel, Hoeben Ann, De Ruysscher Dirk, Vooijs Marc. Patient-Derived Cancer Organoids as Predictors of Treatment Response. Frontiers in Oncology, 11, 2021, 10.3389/fonc.2021.641980
- Guillen, K.P., Fujita, M., Butterfield, A.J. et al. A human breast cancer-derived xenograft and organoid platform for drug discovery and precision oncology. Nat Cancer 3, 232–250 (2022). https://doi.org/10.1038/s43018-022-00337-6
- Golebiewska, A., Hau, AC., Oudin, A. et al. Patient-derived organoids and orthotopic xenografts of primary and recurrent gliomas represent relevant patient avatars for precision oncology. Acta Neuropathol 140, 919–949 (2020). https://doi.org/10.1007/s00401-020-02226-7
- van Renterghem, A.W.J., van de Haar, J. & Voest, E.E. Functional precision oncology using patient-derived assays: bridging genotype and phenotype. Nat Rev Clin Oncol 20, 305–317 (2023). https://doi.org/10.1038/s41571-023-00745-2
- Letai A, Bhola P, Welm AL. Functional precision oncology: Testing tumors with drugs to identify vulnerabilities and novel combinations. Cancer Cell (2022). https://doi.org/10.1016/j.ccell.2021.12.004
- Genta S, Coburn B, Cescon DW, Spreafico A. Patient-derived cancer models: Valuable platforms for anticancer drug testing. Front Oncol. (2022). https://doi.org/10.3389/fonc.2022.976065
- Lõhmussaar, K., Boretto, M., & Clevers, H. (2020). Human-Derived Model Systems in Gynecological Cancer Research. Trends in cancer, 6(12), 1031–1043. https://doi.org/10.1016/j.trecan.2020.07.007
- Meehan TF, Conte N, Goldstein T, et al. PDX-MI: Minimal Information for Patient-Derived Tumor Xenograft Models. Cancer Res. (2017). https://doi.org/10.1158/0008-5472.CAN-17-0582
Top image by CancerModels.org





Join the FEBS Network today
Joining the FEBS Network’s molecular life sciences community enables you to access special content on the site, present your profile, 'follow' contributors, 'comment' on and 'like' content, post your own content, and set up a tailored email digest for updates.