Competition to revolutionize CryoET
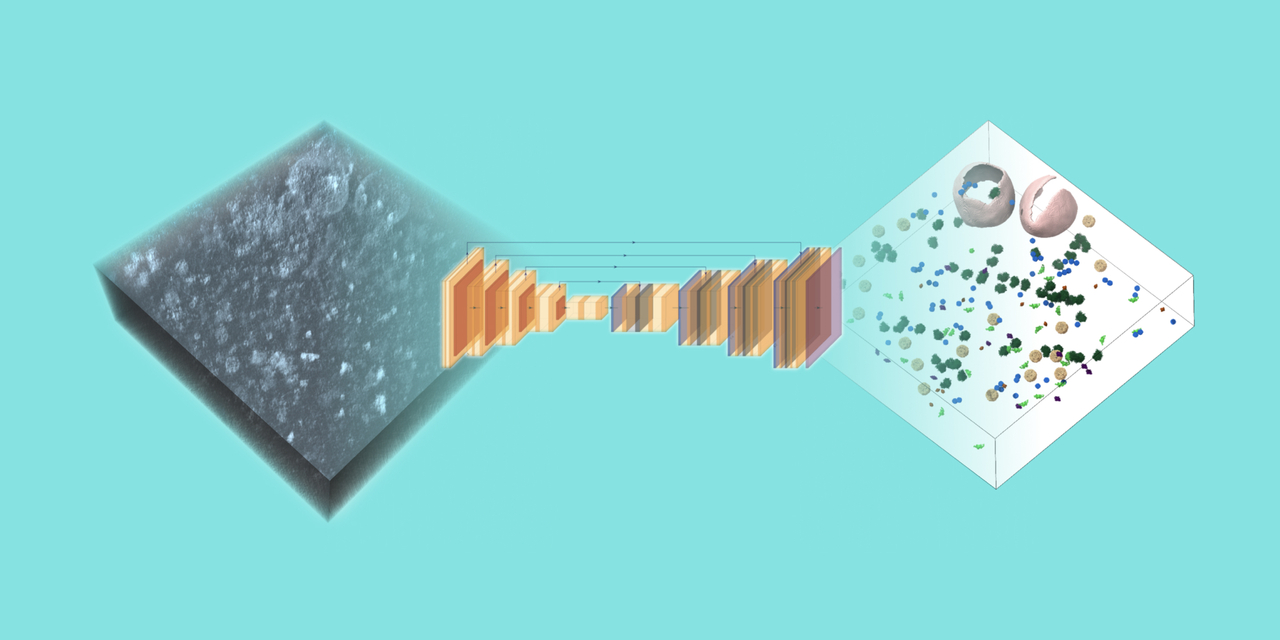
If the 2024 Nobel Prize in Chemistry is any indication, protein structural biology has made great leaps in recent years. Understanding how proteins interact with each other and other parts of the cell, however, remains a key question in biology. Cryo electron tomography (cryoET) is an advanced imaging technology that is poised to answer this question, accelerating discovery of new treatments for disease. Importantly, it’s a technique that can resolve protein complexes in cells to near-atomic level, while preserving the cellular architecture and capturing a wide enough field of view to see the proteins in their biological context.
CryoET can provide thousands of 3D images—called tomograms—in just a few days. But to realize the full potential of this technology, biological objects like proteins and membranes need to be identified in the tomogram through annotation. A given tomogram is typically only about 200 nanometers thick—approximately five hundred times thinner than a sheet of paper—yet packed with information about the structures of the cellular machinery driving health and disease. Manual annotation on a set of tomograms can take months and is near impossible on the total volume of data that already exists, making annotation a major bottleneck in biomedical discovery.
The Chan Zuckerberg Imaging Institute is hosting an international competition to advance our understanding of cell biology by developing machine learning algorithms that can annotate biological particles in 3D images of cells captured by cryoET. These algorithms should be able to perform robust annotation of particles of variable shapes and sizes within the hundreds of 3D images in the competition dataset after being trained on a limited set of available reference annotations from the same dataset. To reduce the onboarding time for competitors, an extensive set of example notebooks is being provided.
Thanks to technological advancements, sample preparation, imaging, image processing, and open access standardized sharing of data have all seen improved efficiency. The field, however, is still limited by this type of annotated data: there is a great and immediate need for machine learning algorithms that can robustly annotate particles of different shapes, sizes and abundances within cells to unlock the discoveries currently trapped in thousands of existing tomograms.
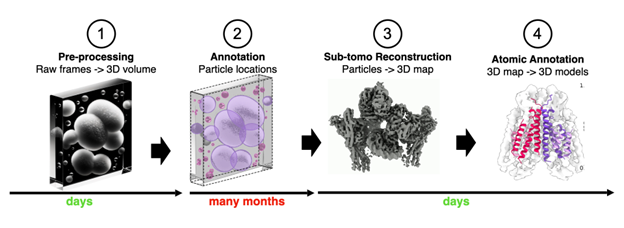
General pipeline from acquisition to creating 3D models from cryoET tomograms. Manual annotation is incredibly time-consuming and can take months for a set of tomograms, representing a significant bottleneck in translating impact from cryoET data.
Annotation strategies that can be readily adapted to datasets acquired under different settings, and that better capture the heterogeneity of biomolecules would be revolutionary compared to currently available approaches.
By design, any tools developed for the purposes of the competition are compatible to be applied to the entire data corpus on the CryoET Data Portal with relative ease, meaning impact will extend beyond the competition. In fact, the CryoET Data Portal was created for this very purpose: so that biologists and developers alike can openly access high-quality, annotated data. All data on the portal is standardized to facilitate the retraining or development of new annotation models and algorithms. All tomograms in the cryoET Data Portal include rich standardized metadata and follow a common data tree structure and common naming conventions. The CZ Imaging Institute is working with developers at EMPIAR to ensure the portal is compatible with their resource, and CZI recently funded a workshop with EMPIAR’s founding group, the European Bioinformatics Institute (EBI), to discuss metadata standards.
There are currently over 15,419 tomograms publicly available in standardized format through the CryoET Data Portal. Only 5%, however, have molecular annotations to date, meaning there is a large pool of standardized data ready to be analyzed with algorithms developed for the competition. This is a unique opportunity because, while there are already machine learning methods applied to cryoET data, they are typically only effective for the specific cases they were developed on and often do not generalize sufficiently to meet the diverse needs of the cryoET community. Even the design of the competition dataset and reference annotations led to the development of several new tools to aid particle picking. This process, however, took several months and substantial manual labor, further underscoring the need for a streamlined and fully automated solution to tomogram annotation.
Machine learning has been successfully leveraged to provide membrane segmentations for over 15,000 of the 3D images currently available in the open access CryoET Data Portal. This was achieved in just 3 days, whereas it would take years to do manually. Annotating particles such as protein complexes, however, is a far more difficult task due to their diversity, lower contrast, and crowding, and remains a critical bottleneck in the cryoET pipeline. Submitted algorithms can also help provide benchmark datasets and standardized pipelines to guide the continuous improvement of machine learning models.
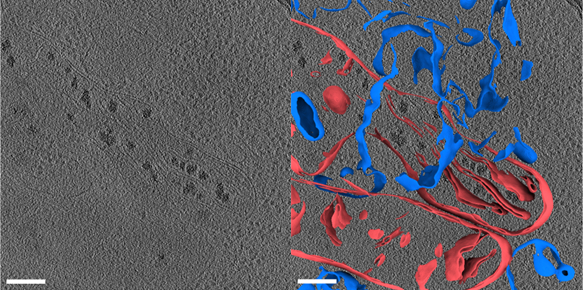
3D tomogram of a human umbilical vein endothelial cell (HUVEC) from Dataset 10176 before (left) and after (right) membrane annotations. Mitochondrial membranes are colored red, other vesicular membranes are colored blue. The scale bar is 200 nm.
The CryoET Data Portal is only a year old, but with a growing number of contributors, collaboration between the cryoET community and machine learning developers can uncover new insights about how all of the components of a cell come together—in different cells and during different states of health, disease and age. We anticipate that well-annotated cryoET data will contribute to building a Virtual Cell Model, revolutionizing our understanding of the structure of the cell in the same way that AlphaFold revolutionized understanding of the structure of proteins. This should ultimately lead to astonishing new insights in biology, and lay the foundation for future medicines.
Enter the competition and encourage your peers to enter as well! For more details, visit: cryoetdataportal.czscience.com/competition
All images by the Chan Zuckerberg Imaging Institute.




Join the FEBS Network today
Joining the FEBS Network’s molecular life sciences community enables you to access special content on the site, present your profile, 'follow' contributors, 'comment' on and 'like' content, post your own content, and set up a tailored email digest for updates.
A wonderful initiative!