My favourite lab technique: Olivier Duss highlights single-molecule fluorescence microscopy

After a PhD at ETH in Zurich, Switzerland, Olivier Duss did a combined postdoc at Scripps Research and Stanford University, USA, studying co-transcriptional ribosome assembly using single-molecule methods. He is now a Group Leader at the Structural and Computational Biology Unit of the European Molecular Biology Laboratory (EMBL), Heidelberg, Germany, where his group works on understanding how RNA interacts with proteins and how protein–RNA complexes assemble and function, using a combination of single-molecule biophysics, structural biology and biochemistry. He received a FEBS Excellence Award in 2022 (funds for equipment/consumables for early-career group leaders).
What do you particularly like about single-molecule fluorescence microscopy?
As the name implies, you can look at individual molecules or cellular machines and you can observe what they are doing. Most biochemical, biophysical and structural techniques need millions of millions of molecules to generate a signal that is large enough to detect. But the information that one obtains from these methods is an average over all the individual molecules. In contrast, being able to observe individual molecules over time, we can see what each one of them is doing. For example, we can observe how individual RNA molecules fold into their active conformation while they are being transcribed by the RNA polymerase or we can track how cellular machines such as the ribosome and the polymerase in bacteria interact and communicate with each other, in real-time while they are doing so. Aptly speaking, these single-molecule experiments allow us to watch a movie of individual molecules in action. This is the most direct way to understand how things work because seeing is believing.
Can you explain the principles of how it works to people not familiar with this?
In order to be able to detect individual molecules, we need to have a very strong signal from each one of them and we need sophisticated equipment to detect these very weak signals. We attach a small but very bright dye – such as a cyanine dye – to each molecule that we want to observe and using fluorescence microscopes, the signal of this single dye becomes detectable. We can use different dyes in order to label different molecules with different colours. This allows us to view several molecules simultaneously and see what they are doing. To observe how different molecules interact with each other, we usually have to attach one molecule of interest to a glass surface. We can do so, for example, by biotinylating both the molecule to be attached and the surface and then using streptavidin to glue the molecule to the surface. By viewing the colours that approach this surface-attached molecule, we know which dye-attached molecules are interacting with our molecule of interest.
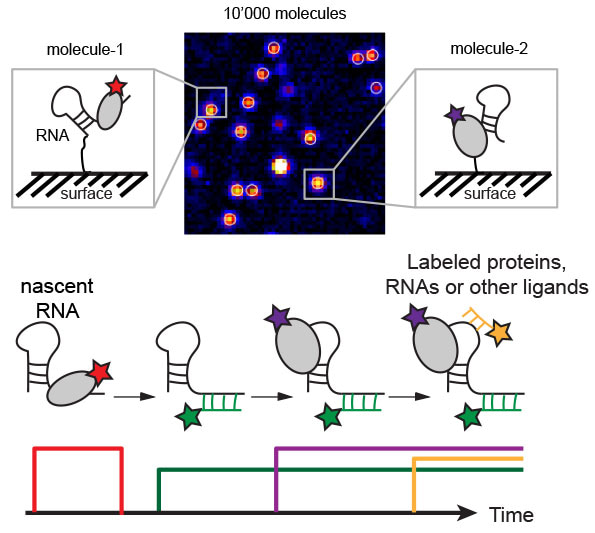
How and when did you start using it and has it taken long to master?
I started using it during my postdoc in the USA. During my PhD, I learned structural biology. While this gives you structural snapshots of molecules at high resolution, most of these methods do not provide dynamic information. Therefore, I wanted to learn a method that allows me to also study how molecules move and interact with each other.
I am still learning. The basic principles are not more complicated than for any other biophysical or structural method. But every biological question to be addressed requires different single-molecule experiments. There are so many possibilities and the fun thing is to think about how to design your next experiment in order to figure out how your biological system works.
What are some of the pitfalls/challenges to avoid?
Because we can detect single molecules, we essentially see everything, also molecules that we are not interested in. Therefore, we need to be very careful when we design experiments, to make sure that what we detect is really what we think it is. We always make several controls and also use complementary biochemical methods to verify that what we see is really what it is.
Are there similar or alternative approaches out there?
There are many different single-molecule techniques, some of which give dynamic information, while some others give other information. For example, a single-molecule technique that recently has gained a lot of attention is cryogenic electron tomography, but it gives structural information of individual molecules only in a static and frozen state. Therefore, combining the information from both techniques, cryogenic electron tomography giving structural snapshots of individual molecules and single-molecule fluorescence microscopy giving dynamic information, allows placing the static snapshots into a dynamic movie, visualizing how single molecules or cellular machines work and interact with each other, in real-time.
What new applications are being seen now?
While single-molecule fluorescence methods usually are limited to two or sometimes three colours and the interaction of two molecules or a structural change within a molecule can be observed, using more colours, up to six, will allow us to look at much more complicated systems. Using reconstitution of complicated biological systems in vitro, we have, for example, been able to simultaneously observe in real-time how an RNA and a protein are being synthesized and, at the same time, how the two cellular machines that produce that RNA and the protein physically interact with each other. We are currently in the process of publishing these new findings. In future, apart from looking at even more cellular processes simultaneously, performing similar experiments also within a living cell may eventually become possible.
Lab webpage: https://www.embl.org/groups/duss/
Suggested reading on single-molecule fluorescence microscopy for newcomers:
Ray. S., Widom, J.R. and Walter, N.G. (2018) Life under the microscope: single-molecule fluorescence highlights the RNA world. Chem. Rev. 118, 8, 4120–4155. https://pubs.acs.org/doi/pdf/10.1021/acs.chemrev.7b00519
Feng, X.A. , Poyton, M.F. and Ha, T. (2021) Multicolor single-molecule FRET for DNA and RNA processes. Curr. Opin. Struct. Biol. 70, 26–33. https://www.sciencedirect.com/science/article/abs/pii/S0959440X21000373
Duss, O., Stepanyuk, G.A., Puglisi, J.D. and Williamson, J.R. (2019) Transient protein-RNA Interactions guide nascent ribosomal RNA folding. Cell 179, 1357–1369.e16. https://doi.org/10.1016/j.cell.2019.10.035
Gor, K. and Duss, O. (2023) Emerging quantitative biochemical, structural, and biophysical methods for studying ribosome and protein–RNA complex assembly. Biomolecules 13, 866. https://doi.org/10.3390/biom13050866
Top image of post: by Gerd Altmann from Pixabay





Join the FEBS Network today
Joining the FEBS Network’s molecular life sciences community enables you to access special content on the site, present your profile, 'follow' contributors, 'comment' on and 'like' content, post your own content, and set up a tailored email digest for updates.