My favourite lab technique: Alessandro Bertero highlights genome editing in human pluripotent stem cells

Alessandro Bertero is an Associate Professor of Experimental Biology in the Dept. of Molecular Biotechnology & Health Sciences of the University of Torino, Italy. After a PhD from the University of Cambridge, UK, on TGF-β signalling in early differentiation of human pluripotent stem cells, he worked as a postdoc then junior faculty member at the University of Washington, Seattle, USA, on 3D chromatin organization dynamics during human cardiogenesis and in dilated cardiomyopathy. In 2021 Alessandro received the Armenise-Harvard Career Development Award to establish his laboratory in Italy. He received a FEBS Excellence Award in 2022 (funds for equipment/consumables for early-career group leaders).
What do you particularly like about genome editing in human pluripotent stem cells?
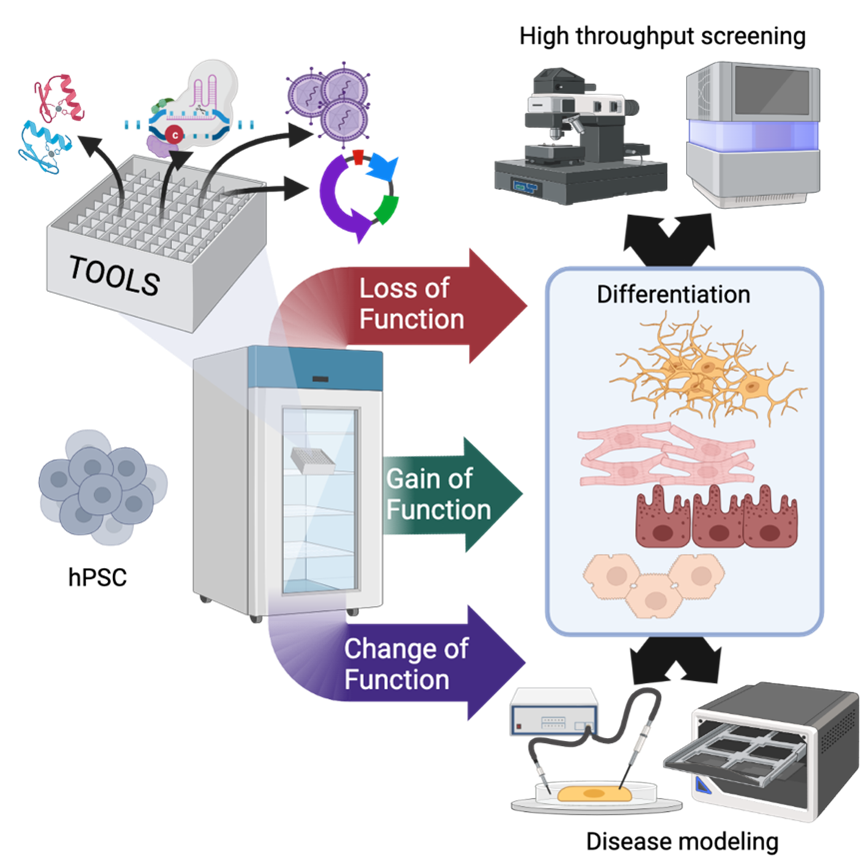
The wide variety of applications it unlocks: by joining two transformative, Nobel prize-worthy technologies, the sky is the limit as far as what one can achieve. Human pluripotent stem cells (hPSCs), particularly those induced by reprogramming of somatic cells (hiPSCs), offer unprecedented opportunities to study rare, transitory or difficult to obtain cell types, and they have opened a new era in regenerative medicine. Genome editing, particularly CRISPR/Cas9, allows us to rewrite sections of the code of life so as to either study gene function or even engineer novel activities in cells. These two methods are incredibly synergistic, and we are beginning to see their impact from labs to bedsides.
Can you explain the principles of how it works to people not familiar with this?
hiPSCs are obtained by expressing a set of embryonic transcription factors in adult cells obtained from a small skin biopsy, a blood draw, or even urine. In the right culture conditions, cells are reprogrammed to a state that mimics that of embryonic stem cells to all intents and purposes, but does not pose the same ethical dilemmas. Plus, hiPSCs can be derived virtually from any individual, which allows us to study genetic diseases and the development of personalized medicines. CRISPR/Cas9 and its variants rely on a small RNA to deploy specific proteins at a predetermined place in the DNA: these can be molecular scissors that can either destroy a gene to inactivate it or facilitate recombination of an exogenous sequence; transcriptional activators or repressors that can up- or down-regulate gene expression; or enzymes that modify one or a few nucleotides to introduce specific mutations.
Combining these two approaches has some nuances, for instance hiPSCs are very sensitive to DNA damage, but can be done quite effectively allowing all kinds of modifications in hiPSCs, such as the correction (or introduction) of gene mutations, the expression of fluorescent reporters, or even the creation of complex synthetic circuits to direct hiPSC differentiation into specific lineages.
How and when did you start using it?
I did my first genome edit over ten years ago at the start of my PhD in Ludovic Vallier’s lab at the University of Cambridge. At the time we worked on human embryonic stem cells and used the “grandfather” of CRISPR/Cas9 (zinc finger nucleases, which are a lot harder to engineer), but even back then I was amazed by what could be achieved. Together with some talented colleagues I developed methods that insert two sets of “instructions” in different genomic regions called genomic safe harbors, as they can be safely edited without affecting cell function and are stably expressed after hPSC differentiation. We applied this approach to either silence [1] or activate [2] gene expression; the latter allowed us to force the differentiation of hPSCs into neurons or skeletal muscle cells by overexpressing master regulators of these cell types. This started as a pure academic project but turned out to have strong industrial potential (empowering both UK-based bit.bio and Netherlands-based Meatable), which showcases the power of genome editing in hPSCs.
What have you learned since then?
That we were blessed by beginners’ luck and that things do not always work as designed on paper. For example, hPSCs are very, very good at identifying and silencing foreign sequences, particularly those of viral or bacterial origin: this can waste months of work as transgenes fail to express reproducibly and/or do not work after hPSC differentiation. Another thing to look out for is the incredible resilience of the human genome to modifications that should knockout a gene: alternative isoforms and genetic compensation can completely rescue an otherwise strong phenotype. There are ways to mitigate both of these risks, but unfortunately these kind of difficulties and solutions are rarely discussed in the literature, which has a bias for positive results. This is why we recently wrote an extensive review to summarize all that we have learned during the last decade: the do’s and don’ts in this arena [3]. We hope this resource will help those approaching the field avoid major pitfalls and reach their scientific objectives more quickly.
Tell us about one of your research papers using this approach
During my postdoc and junior faculty years I deployed genome editing in hPSCs in a very different direction: to increase the safety of cells used for cardiac regeneration [4]. I was working in Chuck Murry’s lab at the University of Washington in Seattle, and we were challenged by transitory yet life-threatening arrythmias induced by the transplantation of cardiomyocytes differentiated from hPSCs in preclinical models of myocardial infarction. The lab’s major hypothesis was that these arrythmias resulted from the immaturity of in vitro differentiated cells and in particular a phenomenon called automaticity (i.e. the spontaneous generation of action potentials), causing an electrical mismatch with the host. My colleagues had been trying all kinds of approaches to mature the cells (i.e. hormones, mechanical stimulation, metabolic substrates) but none seemed to be sufficient. We decided to bring in the big guns, using CRISPR/Cas9 to eliminate the expression of foetal ion channels and overexpress adult ion channels (selected based on a transcriptomic analysis of maturation after cell engraftment). Even this approach proved challenging, but after some five years and a dozen of genome edits we found a combination of three knockout and one knockin that resulted in cardiomyocytes that are virtually quiescent but still able to respond normally to an external electrical stimulus. When transplanted, these cells did not lead to severe arrythmias, and beat in sync with the host.
What new applications are on the horizon?
The emergence of more and more refined genome-edited, hPSC-derived organoid models is enabling the study of normal and abnormal human development in unprecedented detail. Moreover, the combination of another transformative technology, single-cell genomics, has revolutionized the granularity of such studies, as well as the rapid screening of many perturbations within the same experiment. The combination of cell and gene therapy is another exciting avenue. For instance, several groups and companies are developing non-immunogenic hPSCs, and others are engineering them to deliver therapeutic proteins. Last but not least, I am also very excited about the application of genome editing in PSCs of livestock animals, which is emerging as powerful approach to cut down the costs of producing the enormous amounts of cells needed for the field of cellular agriculture (i.e. the generation of meat and seafood directly from cells) to become viable at large scale. This could help mitigate the sustainability challenges faced by our rapidly warming world. As I often say to my students, stem cells and genome editing can improve human life on earth from a holistic perspective: cradle to table and all the way to rocking chair.
Lab webpage: https://www.berterolab.com/
References and suggested reading for newcomers:
1 Bertero, A. et al. (2016) Optimized inducible shRNA and CRISPR/Cas9 platforms for in vitro studies of human development using hPSCs. Development 143, 4405–4418. https://doi.org/10.1242/dev.138081
2 Pawlowski, M. et al. (2017) Inducible and deterministic forward programming of human pluripotent stem cells into neurons, skeletal myocytes, and oligodendrocytes. Stem Cell Rep 8, 803–812. https://doi.org/10.1016/j.stemcr.2017.02.016
3 Balmas, E. et al. (2023) Manipulating and studying gene function in human pluripotent stem cell models. FEBS Lett. https://doi.org/10.1002/1873-3468.14709
4 Marchiano, S. et al. (2023) Gene editing to prevent ventricular arrhythmias associated with cardiomyocyte cell therapy. Cell Stem Cell 30, 396–414. https://doi.org/10.1016/j.stem.2023.03.010
Top image of post: by Pete Linforth from Pixabay.




Join the FEBS Network today
Joining the FEBS Network’s molecular life sciences community enables you to access special content on the site, present your profile, 'follow' contributors, 'comment' on and 'like' content, post your own content, and set up a tailored email digest for updates.