Music & Mutation

When we both turned up to a Manchester Science Festival mixer session at the Museum of Science & Industry, little did we know we would find each other and embark on a sci-art collaboration. We were from different institutions, although just down the road from each other: the Manchester Institute of Biotechnology and the Royal Northern College of Music – what could we possibly work together on? However, shortly after being introduced to each other we had already decided on a project – composing music from DNA sequences. This had of course been done before, but we wanted to put a different spin on it.
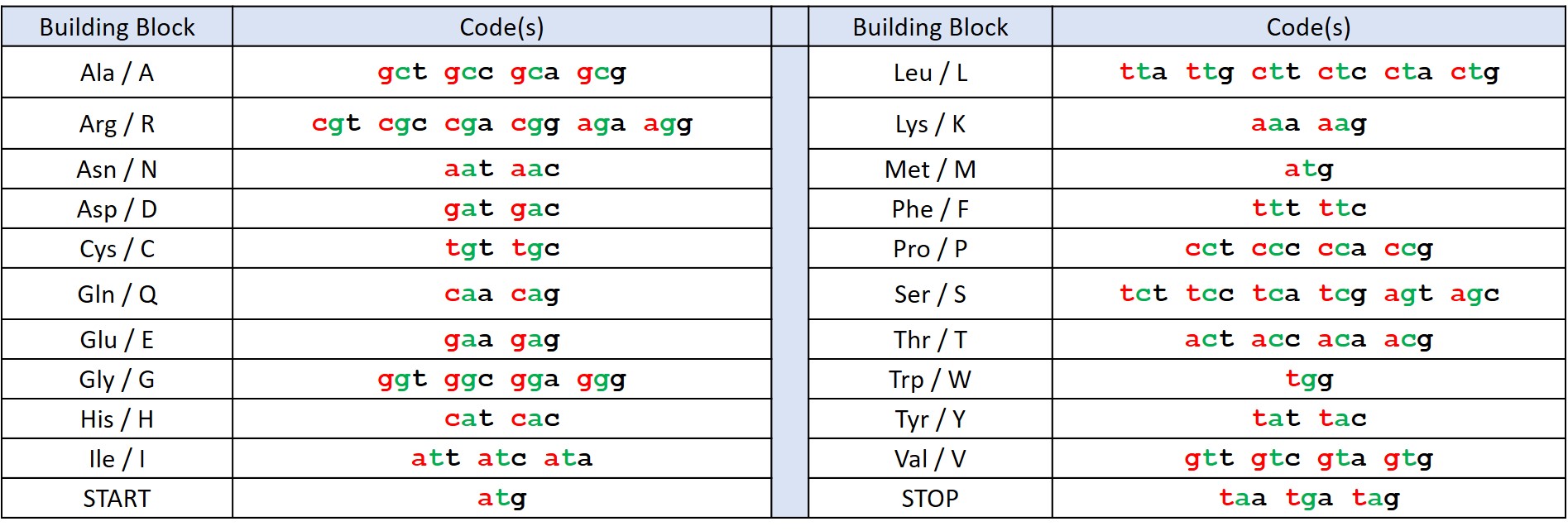
Figure 1: The standard genetic code with each base coloured according to its position in the codon (1st = red, 2nd = green, 3rd = black).
The concept that we wanted to introduce members of the general public to was heterologous expression of useful genes in a laboratory strain organism and why codon optimisation was often needed for this. Put simply, why do we take genes from one organism and put them into another to study in the lab and why do we sometimes have to alter the sequence before doing this. For this we would need something more complex than a representation of each base in musical form – it would be necessary to have a different sound for each codon.
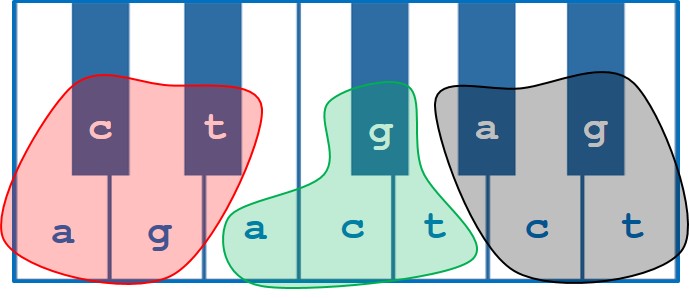
Figure 2: The standard 12 tones of Western music theory represented on a keyboard overlaid with the 12 positional combinations of each DNA base in a given codon – a / c / g / t at the 1st position (red), a / c / g / t at the 2nd position (green) and a / c / g / t at the 3rd position (black).
This was achieved by taking the 12 possible combinations of 4 bases at any one of 3 positions in a codon and representing each by one of the degrees of a standard 12 tone chromatic scale. From this, a suite of 64 distinct chords could be achieved representing each of the 64 codons in the genetic code. Excellent! Now we had a way of turning any DNA sequence into a series of chord progressions that would sound different for any protein coding-gene. The next issue was how to show differences in expression between different organisms.
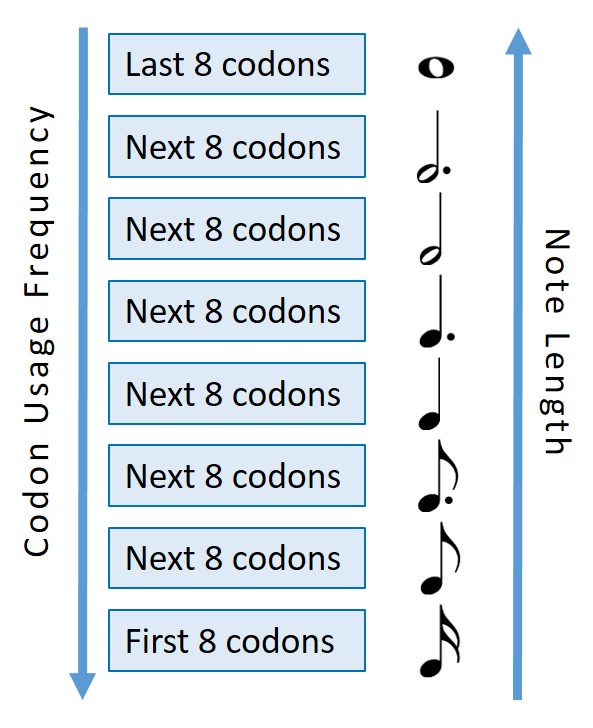
Figure 3: A depiction of how the chords of the associated codons were split into octiles and distributed between 8 different note lengths (increasing note length with decreasing codon usage frequency).
One of the main differences between organisms expressing genes, or at least the main difference when talking about heterologous expression of the same DNA sequence, is the codon usage bias of the organism. Some organisms favour certain codons over others, particularly those that code for the same amino acid, as seen by comparing the frequency of the codons in the genomes of those organisms. To demonstrate this we used 8 different chord lengths and correlated this with the codon usage bias of each codon. For a given organism the 8 least frequent codons where assigned the longest chord length (a semibreve), the second 8 least frequent the next shortest chord length (a dotted minim), the third 8 least frequent the next shortest chord length (a minim), etc. This meant that the same gene sequence could be ‘played’ by different organisms and, although the chords would be in the same order, their relative lengths would vary.

Figure 4: An excerpt of the gene sequence for DdPAL – a phenylalanine ammonia lyase enzyme – as ‘played’ by its host organism Dictyostelium discoideum.
Finally to put our system to work we used some research done at the Manchester Institute of Biotechnology that involved the production of a slime mould enzyme in Escherichia coli using a codon optimised gene sequence. As a full demonstration, 3 ‘movements’ of music for string trio were made: 1) with chords based on the wild-type sequence and lengths based on the host organism’s codon usage bias, 2) with chords based on the wild-type sequence and lengths based on the laboratory strain organism’s codon usage bias, 3) with chords based on the codon optimised sequence and lengths based on the laboratory strain organism’s codon usage bias. These pieces have been performed live and presented in recorded form to members of the general public across a range of events since 2018. What is always striking to us is how different people say the movements sound, even ascribing emotions and colours to them (despite their being no creative licence behind their composition other than the arbitrary chord and note length choices).

Figure 5: Pictures of Nick and Michelle at various events where members of the public were introduced to the Music & Mutation pieces.
This project has shown how concepts that are difficult to explain (even to university-level science students) can be communicated to members of the general public through music. The method gives an extra layer of intellectual accessibility to aid understanding and communicate the techniques used widely in biomolecular science and research. It would be fun to see who else’s research and gene sequences could be turned into pieces of music and communicated to non-specialist audiences!
Further Reading:
More on the science (original DNA sequences): Weise, N.J., Ahmed, S.T., Parmeggiani, F., Galman, J.L., et al. Zymophore identification enables the discovery of novel phenylalanine ammonia lyase enzymes. Sci Rep 7, 13691 (2017). https://doi.org/10.1038/s41598-017-13990-0
More on the music (a sonification concert): Martin, E.J., Knotts, S., Phillips, M., Weise, N.J., et al. Innovative communication of molecular evolution through sound: a biological sonification concert. Evo Edu Outreach 17, 6 (2024). https://doi.org/10.1186/s12052-024-00200-5
All images by Nicholas Weise and The Royal Northern College of Music.




Join the FEBS Network today
Joining the FEBS Network’s molecular life sciences community enables you to access special content on the site, present your profile, 'follow' contributors, 'comment' on and 'like' content, post your own content, and set up a tailored email digest for updates.
This is really cool - what a great collaboration.
Any genes you work with that could be represented in this way? It's really interesting when you hear the music, particularly the stopping and starting (even in wild-type sequences, which I expected would be really smooth...)
Thanks for your reply. I wonder how the sounds change when different genetic codes are used? I often need to consider coding of mitochondrial encoded genes which uses a slightly different code from eukaryotic nuclear code.