AlphaFold five years on: What this new era means for Molecular Life Sciences
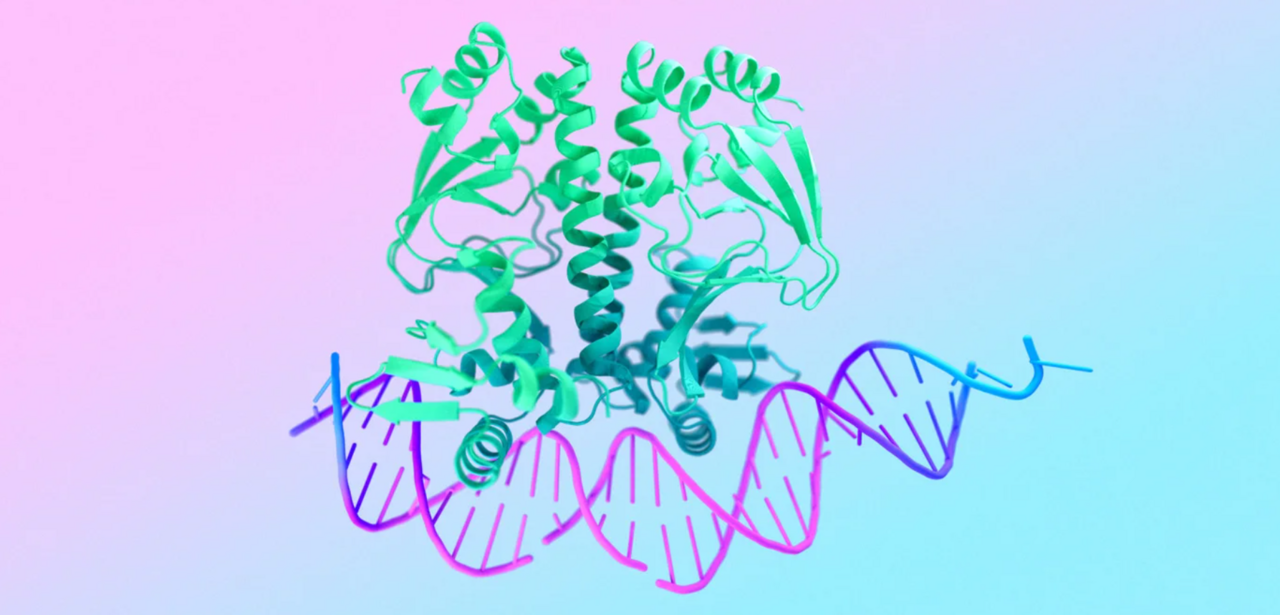
This post offers a brief recap of Google’s (DeepMind) new article on AlphaFold. You can read the full original piece here: https://deepmind.google/blog/alphafold-five-years-of-impact/
Five years ago, AlphaFold 2 demonstrated that AI could predict protein structures with unprecedented accuracy, reshaping one of biology’s oldest challenges. Since then, its influence has spread across many areas of molecular life sciences.
When Google and EMBL-EBI launched the AlphaFold Protein Structure Database in 2021, it quickly became a globally adopted resource. Today it contains 200 million+ predicted structures, freely accessible and used by over 3 million researchers from more than 190 countries. In 2024, this work was recognised with the Nobel Prize in Chemistry.
Impact highlighted in Google’s article
The piece outlines a broad range of ways in which AlphaFold is already helping science:
-
Understanding key biological systems
From bee immunity proteins to the elusive apoB-100 involved in LDL particles, AlphaFold predictions are supporting research that was previously held back by structural uncertainty. -
Supporting discovery and innovation
Over 35,000 publications now cite AlphaFold, and labs using it show increased rates of reporting new experimental structures — suggesting it is helping researchers explore areas that were harder to access experimentally. -
Making structural biology more accessible
Students and early-career scientists have used AlphaFold to learn and publish, even without previous structural biology training — something unthinkable a decade ago.
Looking ahead
Google also describes AlphaFold 3, which extends predictions to DNA, RNA, ligands and entire molecular assemblies, and related models (such as AlphaMissense or AlphaGenome) aimed at understanding disease-associated mutations or designing new protein binders.
These developments hint at a future where digital models increasingly complement laboratory work and help researchers generate and test ideas faster — across many fields, not just structural biology.
Opening the discussion to the FEBS community
This is where your experience matters.
Whether you use AlphaFold daily, occasionally, or simply follow its evolution:
-
How has AlphaFold influenced your work or your thinking?
-
Do you find its predictions reliable for your field?
-
What still feels missing?
-
How do you see tools like AlphaFold fitting into future biochemical research?
💬 Feel free to share thoughts, examples or concerns — all perspectives are welcome!




Join the FEBS Network today
Joining the FEBS Network’s molecular life sciences community enables you to access special content on the site, present your profile, 'follow' contributors, 'comment' on and 'like' content, post your own content, and set up a tailored email digest for updates.